Software tutorial at IFT: Difference between revisions
| Line 370: | Line 370: | ||
and ROOT will run the script <code>Load.C</code> which loads all code and starts the interpreter. From here it is possible to directly run scripts as defined in the <code>Analysis.C</code> file: | and ROOT will run the script <code>Load.C</code> which loads all code and starts the interpreter. From here it is possible to directly run scripts as defined in the <code>Analysis.C</code> file: | ||
ROOT [1] drawBraggPeakGraphFit(...) | ROOT [1] drawBraggPeakGraphFit(...) | ||
'''DISCLAIMER: Some of the materials have been copied from the GATE v7.2 User's guide: http://wiki.opengatecollaboration.org/index.php/Users_Guide_V7.2''' | '''DISCLAIMER: Some of the materials have been copied from the GATE v7.2 User's guide: http://wiki.opengatecollaboration.org/index.php/Users_Guide_V7.2''' | ||
Revision as of 09:50, 16 March 2017
Introduction and overview
This page is meant as a recipe for the software day at IFT, March 20 2017. We have decided that this should take place on Monday, March 20 between 09.00 am and 3.00 pm at the Department of Physics and Technology (our usual meeting room in the 5th floor).
There are certain steps you need to take prior to the meeting. We do not wish to loose time on installation and configuration of the software needed. Thus, it is imperative that you come with your laptops which already have the following installed and configured properly:
Agenda for the day is as follows:
- An introduction to GATE macros, i.e. GATE input scripts
- Setting up a simple simulation geometry in GATE using a proton bencil beam and a water phantom
- Running short simulations
- Examination of the GATE-output files
We think that the above mentioned mini introduction to GATE should take no longer than 1 – 1.5 hours. Rest of the day, we will focus on a more in-depth review of the analysis code written by Helge P.
- Setting up a tracking calorimeter geometry in GATE
- Running short simulations with the detector geometry
- Using the results of the MC simulations, a short «hands-on» introduction to Helge P.’s analysis code written in the Root framework
- A review of all the different modules in the above mentioned analysis code
The final goals of the day will be:
- Setting up a GATE simulation of an example tracking calorimeter including geometry, material specifications and proton beam definition
- Being able to work with the GATE output files (identifying primary protons, secondary particles, calculating deposited dose etc…)
- Being able to run a complete analysis using the Root-analysis code written by Helge P.
As always, check the User guide and tutorial for the DTC Toolkit to find a Wiki-friendly guide.
GATE
Simulations of Preclinical and Clinical Scans in Emission Tomography, Transmission Tomography and Radiation Therapy
Geant4 is a C++ library, where an application / simulation is built by writing certain C++ classes (geometry, beam, scoring, output, physics), and compiling the binaries from where the simulations are run. Only certain modifications to the simulations can be made with the binaries, such as beam settings, certain physics settings as well as geometry objects pre-defined to be variable.
GATE is an application written for Geant4. It was originally meant for PET and SPECT uses, however it is very flexible so many different kinds of detectors can be designed. To run GATE, only macro files written in the Geant4 scripting language (with some GATE specific commands) are needed to build the geometry, scoring, physics and beam. The output is also defined in the macro files, either to ASCII files or to ROOT files.
In each simulation, the user has to:
- define the scanner geometry
- set up the physics processes
- initialize the simulation
- set up the detector model
- define the source(s)
- specify the data output format
- start the acquisition
Introduction to GATE macros
Gate, just as GEANT4, is a program in which the user interface is based on scripts. To perform actions, the user must either enter commands in interactive mode, or build up macro files containing an ordered collection of commands.
Each command performs a particular function, and may require one or more parameters. The Gate commands are organized following a tree structure, with respect to the function they represent. For example, all geometry-control commands start with geometry, and they will all be found under the /geometry/ branch of the tree structure.
When Gate is run, the Idle> prompt appears. At this stage the command interpreter is active; i.e. all the Gate commands entered will be interpreted and processed on-line. All functions in Gate can be accessed to using command lines. The geometry of the system, the description of the radioactive source(s), the physical interactions considered, etc., can be parameterized using command lines, which are translated to the Gate kernel by the command interpreter. In this way, the simulation is defined one step at a time, and the actual construction of the geometry and definition of the simulation can be seen on-line. If the effect is not as expected, the user can decide to re-adjust the desired parameter by re-entering the appropriate command on-line. Although entering commands step by step can be useful when the user is experimenting with the software or when he/she is not sure how to construct the geometry, there remains a need for storing the set of commands that led to a successful simulation.
Macros are ASCII files (with '.mac' extension) in which each line contains a command or a comment. Commands are GEANT4 or Gate scripted commands; comments start with the character ' #'. Macros can be executed from within the command interpreter in Gate, or by passing it as a command-line parameter to Gate, or by calling it from another macro. A macro or set of macros must include all commands describing the different components of a simulation in the right order. Usually these components are visualization, definitions of volumes (geometry), systems, digitizer, physics, initialization, source, output and start. These steps are described in the next sections. A single simulation may be split into several macros, for instance one for the geometry, one for the physics, etc. Usually, there is a master macro which calls the more specific macros. Splitting macros allows the user to re-use one or more of these macros in several other simulations, and/or to organize the set of all commands. To execute a macro (mymacro.mac in this example) from the Linux prompt, just type :
Gate mymacro.mac
To execute a macro from inside the Gate environment, type after the "Idle>" prompt:
Idle>/control/execute mymacro.mac
And finally, to execute a macro from inside another macro, simply write in the master macro:
/control/execute mymacro.mac
Setting up a simple simulation geometry in GATE using a pencil beam and a water phantom
Visualization
First we may want to set up a visualization engine to see what's going on. This is optional, and runs in batch mode should not be visualized! Here we use the opengl visualizer OGLX, but different kinds of visualization engines are discussed in the GATE Wiki [[1]]
/vis/open/OGLX /vis/viewer/reset /vis/viewer/set/viewpointThetaPhi 60 60 /vis/viewer/zoom 1 /vis/viewer/set/style surface /vis/drawVolume /tracking/storeTrajectory 1 /vis/scene/endOfEventAction accumulate /vis/viewer/update
Most of these commands are self explainatory. By using the storeTrajectory command, all particles are displayed together with the geometry.
Materials database
The default material assigned to a new volume is Air. The list of available materials is defined in the GateMaterials.db file. It's included in the Gate folder, and should be copied to the active directory. It is easy to add new materials to the file, just have a look at the file.
/gate/geometry/setMaterialDatabase MyMaterialDatabase.db
Geometry
Apart from specialized geometries such as PET, SPECT, CT, the general geometry is called as scanner. It must be placed within the world volume, and all parts of the detector (to be scored) be placed within the scanner volume.
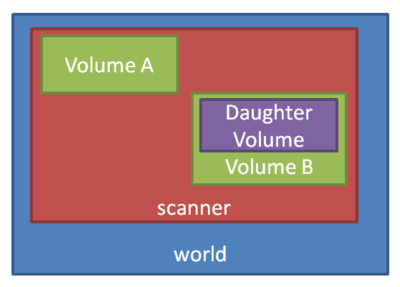
To construct a simple water phantom geometry of 30x30x30 cm, use the following commands:
/gate/world/geometry/setXLength 1000. cm /gate/world/geometry/setYLength 1000. cm /gate/world/geometry/setZLength 1000. cm
So we've defined a world geometry of 1 m3. It must be larger than all its daughter volumes. Let's put the scanner volume inside the world volume. Since it's not already defined (the world volume was), we must insert a box object (with parameters XLength, YLength, ZLength as the side measurements of the box):
/gate/world/daughters/name scanner /gate/world/daughters/insert box /gate/scanner/geometry/setXLength 100. cm /gate/scanner/geometry/setYLength 100. cm /gate/scanner/geometry/setZLength 100. cm /gate/scanner/vis/forceWireframe
Inside this scanner volume (the default material is Air), let's finally put the water phantom (to start at [math]\displaystyle{ z=0 }[/math]):
/gate/scanner/daughters/name phantom /gate/scanner/daughters/insert box /gate/phantom/geometry/setXLength 30. cm /gate/phantom/geometry/setYLength 30. cm /gate/phantom/geometry/setZLength 30. cm /gate/phantom/placement/setTranslation 0 0 -35. cm # - 100/2 + 30/2 /gate/phantom/setMaterial Water /gate/phantom/vis/forceWireframe
Sensitive Detectors
The scoring system in Geant4/GATE is based around Sensitive Detectors (SD). If a volume is a daughter volume (or granddaughter, ...), it may be assigned as a SD. This process is super simple in GATE:
/gate/phantom/attachCrystalSD
Physics
There are many physics lists to choose from in Geant4/GATE. For proton therapy and detector simulations, I most often use a combination of a low-energy-friendly hadronic list and the variable-steplength (for Bragg Peak accuracy) electromagnetic list. From the Geant4 reference physics webpage [[2]]:
- QGSP: QGSP is the basic physics list applying the quark gluon string model for high energy interactions of protons, neutrons, pions, and Kaons and nuclei. The high energy interaction creates an exited nucleus, which is passed to the precompound model modeling the nuclear de-excitation.
- QGSP_BIC: Like QGSP, but using Geant4 Binary cascade for primary protons and neutrons with energies below ~10GeV, thus replacing the use of the LEP model for protons and neutrons In comparison to teh LEP model, Binary cascade better describes production of secondary particles produced in interactions of protons and neutrons with nuclei.
- emstandard_opt3 designed for any applications required higher accuracy of electrons, hadrons and ion tracking without magnetic field. It is used in extended electromagnetic examples and in the QGSP_BIC_EMY reference Physics List. The corresponding physics
The physics list to use all of these is called QGSP_BIC_EMY. It is loaded with the command
/gate/physics/addPhysicsList QGSP_BIC_EMY
In addition, in order to accurately represent the water in the water phantom, we define the current recommended value for the mean ionization potential for water, which is [math]\displaystyle{ 75\ \mathrm{eV} }[/math]. This can be performed for all materials, and it will override Bragg's additivity rule.
/gate/geometry/setIonisationPotential Water 75 eV
Initialization
After the geometry and physics has been set, initialize the run!
/gate/run/initialize
Proton beam
/gate/source/addSource PBS PencilBeam /gate/source/PBS/setParticleType proton /gate/source/PBS/setEnergy 188.0 MeV /gate/source/PBS/setSigmaEnergy 1.0 MeV /gate/source/PBS/setPosition 0 0 -10. mm /gate/source/PBS/setSigmaX 2 mm /gate/source/PBS/setSigmaY 4 mm /gate/source/PBS/setSigmaTheta 3.3 mrad /gate/source/PBS/setSigmaPhi 3.8 mrad /gate/source/PBS/setEllipseXThetaEmittance 15 mm*mrad /gate/source/PBS/setEllipseXThetaRotationNorm negative /gate/source/PBS/setEllipseYPhiEmittance 20 mm*mrad /gate/source/PBS/setEllipseYPhiRotationNorm negative /gate/application/setTotalNumberOfPrimaries 5000
It is tricky to use this beam since all parameters need to match, so an alternative is to use a uniform General Particle Source:
/gate/source/addSource uniformBeam gps /gate/source/uniformBeam/gps/particle proton /gate/source/uniformBeam/gps/ene/type Gauss /gate/source/uniformBeam/gps/ene/mono 188 MeV /gate/source/uniformBeam/gps/ene/sigma 1 MeV /gate/source/uniformBeam/gps/type Plane /gate/source/uniformBeam/gps/shape Square /gate/source/uniformBeam/gps/direction 0 0 1 /gate/source/uniformBeam/gps/halfx 0 mm /gate/source/uniformBeam/gps/halfy 0 mm /gate/source/uniformBeam/gps/centre 0 0 -1 cm /gate/application/setTotalNumberOfPrimaries 5000
Output
For this tutorial, we will use the ROOT output.
/gate/output/root/enable /gate/output/root/setFileName gate_simulation
Running the simulation
To finalize the macro file, start the randomization engine and run!
/gate/random/setEngineName MersenneTwister /gate/random/setEngineSeed auto /gate/application/start
Running short simulations
To run a simulation, create a macro file with the lines as descibed above, and run it with
$ Gate waterphantom.mac
The terminal output describes the geometry, physics, etc.
It is also possible to use aliases in the macro file. For example, to simplify the energy selection, substitute with the line
/gate/source/PBS/setEnergy {energy} MeV
and run the macro with
$ Gate -a '[energy,175]' waterphantom.mac
Multiple aliases can be stacked:
$ Gate -a '[energy,175] [phantomsize,45]' waterphantom.mac
if you have defined multiple alises in the macro file. It is sadly not possible to do calculations in the macro language, so you have to do that through bash (newvalue=`echo "$oldvalue/2" | bc`).
Examination of the GATE output files
The ROOT output file(s) from the simulation can be opened several ways:
- By using the built-in
TBrowserto look at scoring variable distributions - By using loading the ROOT Tree into a C++ program and looping over events (interactions)
Using the built-in TBrowser
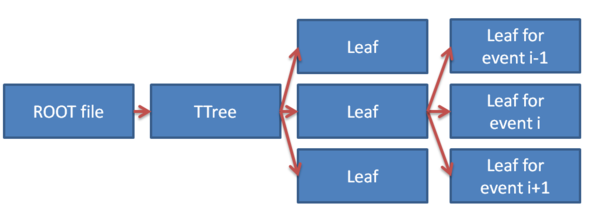
The hierarchy for the files are shown in the image below:
In Gate, the TTree is called Hits, and the leaves are named after the different variables that are automatically scored:
PDGEncoding - The Particle ID
trackID - Track number following a mother particle
parentID - The parent track's event ID. 0 if the current particle is a beam particle
time - Time in simulation (for ToF in PET, etc.)
edep - Deposited energy in this event / interaction
stepLength - The length of the current step
posX - Global X position of event
posY - Global Y position of event
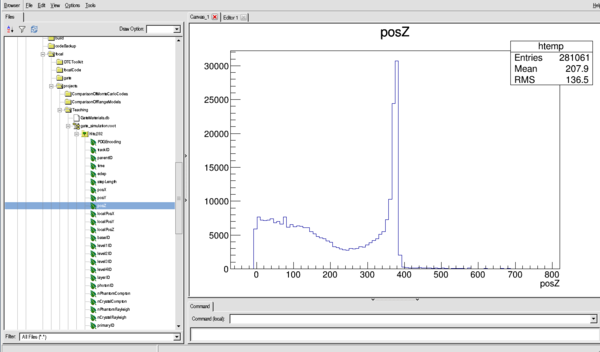
posZ - Global Z position of event
localPosX - Local (in mother volume) X position of event
localPosY - Local (in mother volume) Y position of event
localPosZ - Local (in mother volume) Z position of event
baseID - ID of mother volume scanner, == 0 if only one scanner defined
level1ID - ID of 1st level of volume hierarchy
level2ID - ID of 2nd level of volume hierarchy
level3ID - ID of 3rd level of volume hierarchy
level4ID - ID of 4th level of volume hierarchy
sourcePosX - Global X position of source particle
sourcePosY - Global Y position of source particle
sourcePosZ - Global X position of source particle
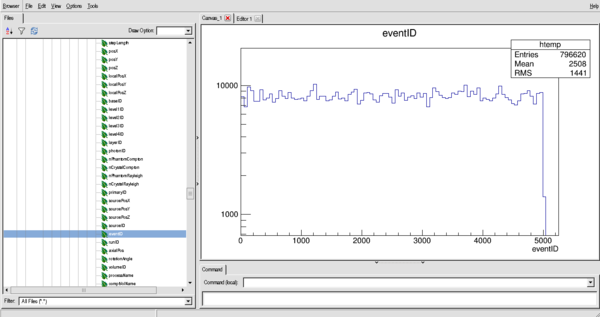
eventID - History number (important!!)
volumeID - ID of current volume (useful to isolate particles in a specific part of a fully scored volume)
processName - A string containing the name of the interaction type:
- hIoni: Ionization by hadron
- Transportation: No special interactions (usually from step limiter)
- eIoni: Ionization by electron
- ProtonInelastic: Inelastic nuclear interaction of proton
- compt: Compton scattering
- ionIoni: Ionization by ion
- msc: Multiple Coulomb Scattering process
- hadElastic: Elastic hadron / proton scattering
An example of the distribution of eventID (in histogram form, this is the number of interactions per particle (if bin size = 1))
$ root ROOT [0] new TBrowser
Or for the Z distribution (see the Bragg Peak)
Opening the files in C++
It is quite simple to open the generated ROOT files in a C++ program.
In openROOTFile.C:
#include <TTree.h>
#include <TFile.h>
using namespace std;
void Run() {
TFile *f = new TFile("gate_simulation.root");
TTree *tree = (TTree*) f->Get("Hits"); // The TTree in the GATE file is called Hits
// Declare the variables (leafs) to be readout
Float_t x,y,z,edep;
Int_t eventID, parentID;
// Make a connection between the declared variables and the leafs
tree->SetBranchAddress("posX", &x);
tree->SetBranchAddress("posY", &y);
tree->SetBranchAddress("posZ", &z);
tree->SetBranchAddress("edep", &edep);
tree->SetBranchAddress("eventID", &eventID);
tree->SetBranchAddress("parentID", &parentID);
// Loop over all the entries in the tree
for (Int_t i=0, i < tree->GetEntries(); ++i) {
tree->GetEntry(i);
if (eventID > 2) break; // To limit the output!
if (parentID != 0) continue; // Only show results from primary particles
printf("Primary particle with event ID %d has an interaction with %.2f MeV energy loss at (x,y,z) = (%.2f, %.2f, %.2f).\n", eventID, edep, x, y, z);
}
delete f;
}
Then you can run the program with
$ root ROOT [0] .L openROOTFile.C+ // The + tells ROOT to compile the code ROOT [1] Run();
Please note that it is also possible to make a complete class to read out the root files using ROOT's MakeClass function. See [[3]].
Test case: Finding the range and straggling of a proton beam
#include <TTree.h>
#include <TH1F.h>
#include <TFile.h>
using namespace std;
void Run() {
TFile * f = new TFile("gate_simulation.root");
TTree * tree = (TTree*) f->Get("Hits"); // The TTree in the GATE file is called Hits
TH1F * rangeHistogram = new TH1F("rangeHistogram", "Stopping position for protons"; 800, 0, 400); // Histogram 1D with Float values
Float_t z;
Int_t eventID, parentID;¨
Int_t lastEventID = -1;
Float_t lastZ = -1;
tree->SetBranchAddress("posZ", &z);
tree->SetBranchAddress("eventID", &eventID);
tree->SetBranchAddress("parentID", &parentID);
for (Int_t i=0, i < tree->GetEntries(); ++i) {
tree->GetEntry(i);
if (parentID != 0) continue;
// Check if this is the first event of a primary particle
if (eventID != lastEventID && lastEventID >= 0) {
rangeHistogram->Fill(lastZ);
}
// Store the current variables
lastZ = z;
lastEventID = eventID;
}
rangeHistogram->Draw();
// Make a Gaussian fit to the range
TF1 * fit = new TF1("fit", "gaus");
rangeHistogram->Fit("fit", "", 350, 400); // Most probable values for fit is in this range, ROOT is quite sensitive to Gaussians occupying only a small part of the histogram, so give narrow fit range
printf("The range of the proton beam is %.3f +- %.3f mm.\n", fit->GetParameter(1), fit->GetParameter(2));
}
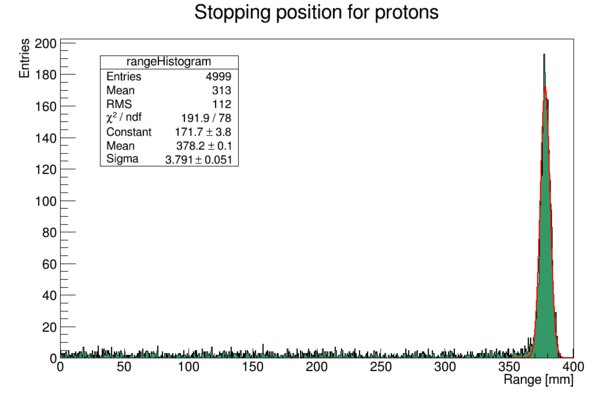
This time, the program will yield the following output (from a 250 MeV beam):
The range of the proton beam is 378.225 mm +- 3.791 mm
With the following histogram (I've added some color and a SetOptFit to the legend)
Review of the analysis code by Helge Pettersen
Setting up a tracking calorimeter geometry in GATE
Running short simulations with the detector geometry
"Hands on" to the analysis code
A review of the different modules in the code
The Digital Tracking Calorimeter Toolkit is located at Helge's github (but should be moved to the Gitlab when ready).
To clone the project, run
git clone https://github.com/HelgeEgil/focal
in a new folder to contain the project. The folder structure will be
DTCToolkit/ <- the reconstruction and analysis code DTCToolkit/Analysis <- User programs for running the code DTCToolkit/Classes <- All the classes needed for the project DTCToolkit/Data <- Data files: Range-energy look up tables, Monte Carlo code, LET data from experiments, the beam data from Groningen, ... DTCToolkit/GlobalConstants <- Constants to adjust how the programs are run. Material parameters, geometry, ... DTCToolkit/HelperFunctions <- Small programs to help running the code. DTCToolkit/OutputFiles <- All output files (csv, jpg, ...) should be put here DTCToolkit/RootFiles <- ROOT specific configuration files. DTCToolkit/Scripts <- Independent scripts for helping the analysis. E.g. to create Range-energy look up tables from Monte Carlo data gate/ <- All Gate-related files gate/python <- The DTC geometry builder projects/ <- Other projects related to WP1
The best way to learn how to use the code is to look at the user programs, e.g. Analysis.C::DrawBraggPeakGraphFit which is the function used to create the Bragg Peak model fits and beam range estimation used in the 2017 NIMA article. From here it is possible to follow what the code does. It is also a good idea to read through what the different classes are and how they interact:
Hit: A (int x,int y,int layer, float edep) object from a pixel hit. edep information only from MCHits: ATClonesArraycollection of Hit objectsCluster: A (float x, float y, int layer, float clustersize) object from a cluster ofHits The (x,y) position is the mean position of all involved hits.Clusters: ATClonesArraycollection ofClusterobjects.Track: ATClonesArraycollection ofClusterobjects... But only one per layer, and is connected through a physical proton track. Many helpful member functions to calculate track properties.Tracks: ATClonesArraycollection ofTrackobjects.Layer: The contents of a single detector layer. Is stored as aTH2Fhistogram, and has aLayer::findHitsfunction to find hits, as well as the cluster diffusion modelLayer::diffuseLayer. It is controlled from aCalorimeterFrameobject.CalorimeterFrame: The collection of allLayers in the detector.DataInterface: The class to talk to DTC data, either through semi-Hitobjects as retrieved from Utrecht from the Groningen beam test, or from ROOT files as generated in Gate.
To run the code, do
[DTCToolkit] $ root Load.C
and ROOT will run the script Load.C which loads all code and starts the interpreter. From here it is possible to directly run scripts as defined in the Analysis.C file:
ROOT [1] drawBraggPeakGraphFit(...)
DISCLAIMER: Some of the materials have been copied from the GATE v7.2 User's guide: http://wiki.opengatecollaboration.org/index.php/Users_Guide_V7.2