Cluster convolution for GATE: Difference between revisions
From pCT
Made page |
No edit summary |
||
| Line 41: | Line 41: | ||
#include <TTree.h> | #include <TTree.h> | ||
#include <TFile.h> | #include <TFile.h> | ||
#include < | #include <TRandom3.h> | ||
#include <list> | #include <list> | ||
#include <vector> | #include <vector> | ||
| Line 62: | Line 62: | ||
Float_t pixel_size = 0.02929; | Float_t pixel_size = 0.02929; | ||
Float_t x,y,z,outX, outY, outZ; | |||
Float_t x,y,z | |||
Int_t parentID, eventID, outEventID, outParentID, nClusters, timeInClockSpeed, PDGEncoding; | Int_t parentID, eventID, outEventID, outParentID, nClusters, timeInClockSpeed, PDGEncoding; | ||
treeSimulationIn->SetBranchAddress("posX",&x); | treeSimulationIn->SetBranchAddress("posX",&x); | ||
treeSimulationIn->SetBranchAddress("posY",&y); | treeSimulationIn->SetBranchAddress("posY",&y); | ||
treeSimulationIn->SetBranchAddress("posZ",&z); | treeSimulationIn->SetBranchAddress("posZ",&z); | ||
treeSimulationIn->SetBranchAddress("eventID",&eventID); | treeSimulationIn->SetBranchAddress("eventID",&eventID); | ||
treeSimulationIn->SetBranchAddress("parentID",&parentID); | treeSimulationIn->SetBranchAddress("parentID",&parentID); | ||
| Line 102: | Line 90: | ||
treeSimulationIn->GetEntry(i); | treeSimulationIn->GetEntry(i); | ||
timeInClockSpeed = 400 * eventID; | timeInClockSpeed = 400 * eventID; // 10 us readout speed | ||
outParentID = parentID; | outParentID = parentID; // =0 for a primary particle, >0 for secondaries | ||
outEventID = eventID; | outEventID = eventID; | ||
outZ = z; | outZ = z; | ||
randomClusterIdx = gRand->Integer(nClusters); | randomClusterIdx = gRand->Integer(nClusters); | ||
| Line 113: | Line 99: | ||
idx_x = 0; | idx_x = 0; | ||
if (abs(PDGEncoding) == 2212) { | if (abs(PDGEncoding) == 2212) { // Only apply this for protons | ||
for (Int_t n : *hit_array) { // loop x | for (Int_t n : *hit_array) { // loop x | ||
for (int binPosPow = 0; binPosPow < 10; binPosPow++) { // loop y | for (int binPosPow = 0; binPosPow < 10; binPosPow++) { // loop y | ||
| Line 121: | Line 107: | ||
outY = y + (binPosPow - y_mean) * pixel_size; | outY = y + (binPosPow - y_mean) * pixel_size; | ||
treeSimulationOut->Fill(); | treeSimulationOut->Fill(); | ||
} | } | ||
} | } | ||
| Line 130: | Line 115: | ||
treeSimulationOut->Write(); | treeSimulationOut->Write(); | ||
} | } | ||
</source> | </source> | ||
Revision as of 14:05, 25 February 2019
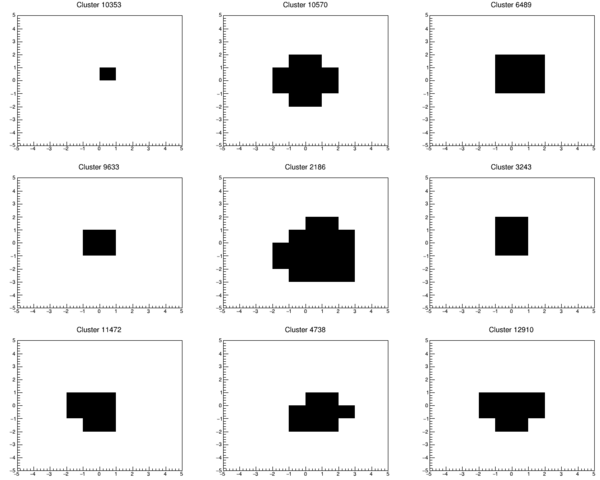
In her MSc project, Silje Grimstad made a filtered library of the clusters from the summer beamtest in Heidelberg. The result is ~20k Helium clusters stored as a x/y array in a ROOT file:
| x_mean | y_mean | size | hit_array | height | width | x_start | y_start | i_event | combined |
|---|---|---|---|---|---|---|---|---|---|
| float | float | int | list<int> | int | int | int | int | int | bool |
A random sample of 9 clusters is shown below:
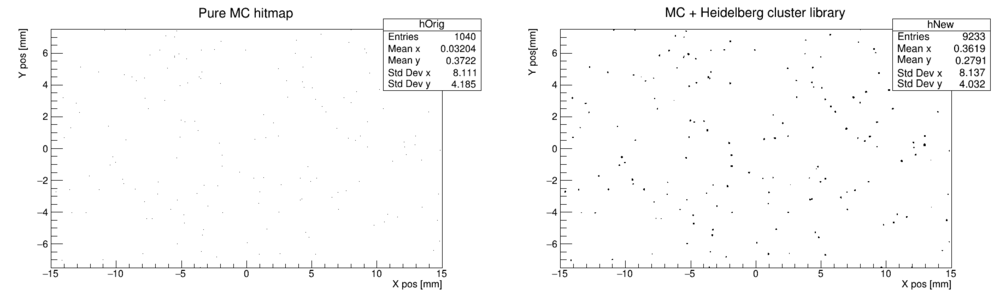
We want to convolute a GATE simulation with clusters from this library. For each hit in the MC data, we pick a random cluster from the library and draw it around the hit position. To do this, use the code at the bottom, with the following result:
The needed cluster database for this is located at GitHib.
#include <TTree.h>
#include <TFile.h>
#include <TRandom3.h>
#include <list>
#include <vector>
using namespace std;
void convert_root_file() {
TFile *fCluster = new TFile("database_combined_clusters_filter.root", "READ");
TFile *fSimulationIn = new TFile("simulationToBeClustered.root", "READ");
TFile *fSimulationOut = new TFile("simulationToBeClustered_clustered.root", "recreate");
TTree *treeCluster = (TTree*) fCluster->Get("database");
TTree *treeSimulationIn = (TTree*) fSimulationIn->Get("Hits");
TTree *treeSimulationOut = new TTree("Hits", "Clustered GATE tree");
Double_t x_mean, y_mean;
Int_t randomClusterIdx, binPos, idx_x;
list<Int_t> *hit_array = 0;
TBranch *b_hit_array;
TRandom3 *gRand = new TRandom3(0);
Float_t pixel_size = 0.02929;
Float_t x,y,z,outX, outY, outZ;
Int_t parentID, eventID, outEventID, outParentID, nClusters, timeInClockSpeed, PDGEncoding;
treeSimulationIn->SetBranchAddress("posX",&x);
treeSimulationIn->SetBranchAddress("posY",&y);
treeSimulationIn->SetBranchAddress("posZ",&z);
treeSimulationIn->SetBranchAddress("eventID",&eventID);
treeSimulationIn->SetBranchAddress("parentID",&parentID);
treeSimulationIn->SetBranchAddress("PDGEncoding", &PDGEncoding);
treeSimulationOut->Branch("posX", &outX, "posX/F");
treeSimulationOut->Branch("posY", &outY, "posY/F");
treeSimulationOut->Branch("posZ", &outZ, "posZ/F");
treeSimulationOut->Branch("eventID", &outEventID, "eventID/I");
treeSimulationOut->Branch("parentID", &outParentID, "parentID/I");
treeSimulationOut->Branch("clockTime", &timeInClockSpeed, "clockTime/I");
treeCluster->SetBranchAddress("x_mean", &x_mean);
treeCluster->SetBranchAddress("y_mean", &y_mean);
treeCluster->SetBranchAddress("hit_array", &hit_array, &b_hit_array);
nClusters = treeCluster->GetEntriesFast();
randomClusterIdx = gRand->Integer(nClusters);
treeCluster->GetEntry(randomClusterIdx);
for (Int_t i=0, N = treeSimulationIn->GetEntries(); i<N; ++i) {
treeSimulationIn->GetEntry(i);
timeInClockSpeed = 400 * eventID; // 10 us readout speed
outParentID = parentID; // =0 for a primary particle, >0 for secondaries
outEventID = eventID;
outZ = z;
randomClusterIdx = gRand->Integer(nClusters);
treeCluster->GetEntry(randomClusterIdx);
idx_x = 0;
if (abs(PDGEncoding) == 2212) { // Only apply this for protons
for (Int_t n : *hit_array) { // loop x
for (int binPosPow = 0; binPosPow < 10; binPosPow++) { // loop y
binPos = pow(2, binPosPow);
if (binPos & n) {
outX = x + (idx_x - x_mean) * pixel_size;
outY = y + (binPosPow - y_mean) * pixel_size;
treeSimulationOut->Fill();
}
}
idx_x++;
}
}
}
treeSimulationOut->Write();
}